%Clustering on COIL20
clear;
load('COIL20.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L1-norm equal to 1
fea = NormalizeFea(fea,1,1);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the original space. MIhat: ',num2str(MIhat)]);
%Clustering in the original space. MIhat: 0.7108
%NMF learning
options = [];
options.kmeansInit = 0;
options.maxIter = 100;
options.nRepeat = 1;
options.alpha = 0;
%when alpha = 0, GNMF boils down to the ordinary NMF.
rand('twister',5489);
[~,V] = GNMF(fea',nClass,[],options); %'
%Clustering in the NMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the NMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the NMF subspace. MIhat: 0.70246
%GNMF learning
options = [];
options.WeightMode = 'Binary';
W = constructW(fea,options);
options.maxIter = 100;
options.nRepeat = 1;
options.alpha = 100;
rand('twister',5489);
[~,V] = GNMF(fea',nClass,W,options); %'
%Clustering in the GNMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the GNMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the GNMF subspace. MIhat: 0.8399
%SDNMF learning
options = [];
options.WeightMode = 'Binary';
W = constructW(fea,options);
options.maxIter = 100;
options.lambda = 100;
options.gamma = 10;
options.alpha = 10;
rand('twister',5489);
[~,V] = SDNMF(fea',nClass,W,options);
%Clustering in the SDNMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the SDNMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the SDNMF subspace. MIhat: 0.8559
Code download
See more examples on incremental active learning using MAED
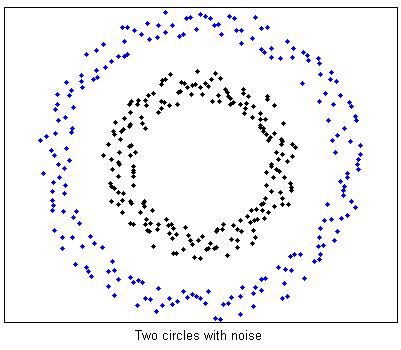
%Two circles data with noise
clear;
rand('twister',5489);
[fea, gnd] = GenTwoNoisyCircle();
split = gnd ==1;
figure(1);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
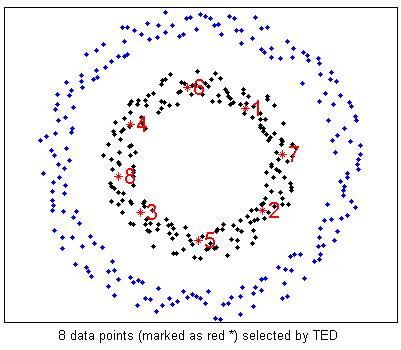
%Actively select 8 examples using TED
options = [];
options.KernelType = 'Gaussian';
options.t = 0.5;
options.ReguBeta = 0;
%MAED boils down to TED when ReguBeta = 0;
smpRank = MAED(fea,8,options);
figure(2);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
hold on;
for i = 1:length(smpRank)
plot(fea(smpRank(i),1),fea(smpRank(i),2),'*r');
text(fea(smpRank(i),1),fea(smpRank(i),2),['\fontsize{16} \color{red}',num2str(i)]);
end
hold off;
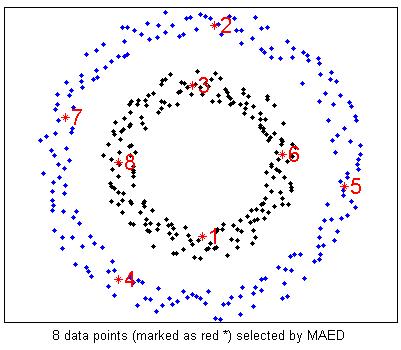
%Actively select 8 examples using MAED
options = [];
options.KernelType = 'Gaussian';
options.t = 0.5;
options.ReguBeta = 100;
smpRank = MAED(fea,8,options);
figure(3);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
hold on;
for i = 1:length(smpRank)
plot(fea(smpRank(i),1),fea(smpRank(i),2),'*r');
text(fea(smpRank(i),1),fea(smpRank(i),2),['\fontsize{16} \color{red}',num2str(i)]);
end
hold off;
Code download
See more examples on out-of-sample retrieval using EMR
%Ranking on the USPS data set (9298 samples with 256 dimensions)
clear;
load('USPS.mat');
gnd = gnd - 1;
%Pick a query point with digit '2'
digit = 2;
idx = find(gnd == digit);
queryIdx = idx(10);
nSmp = size(fea,1);
y0 = zeros(nSmp,1);
y0(queryIdx) = 1;
%Ranking with Euclidean distance
D = EuDist2(fea(queryIdx,:),fea);
[dump,idx]=sort(D);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(fea(queryIdx,:),[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);
%Ranking with Manifold Ranking
tic;
rand('twister',5489);
Woptions.k = 5;
W = constructW(fea, Woptions);
D_mhalf = full(sum(W,2).^-.5);
D_mhalf = spdiags(D_mhalf,0,nSmp,nSmp);
S = D_mhalf*W*D_mhalf;
alpha = 0.99;
S = speye(nSmp)-alpha*S;
y = S\y0;
toc;
%Elapsed time is 2.247750 seconds.
[dump,idx]=sort(-y);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(fea(queryIdx,:),[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);
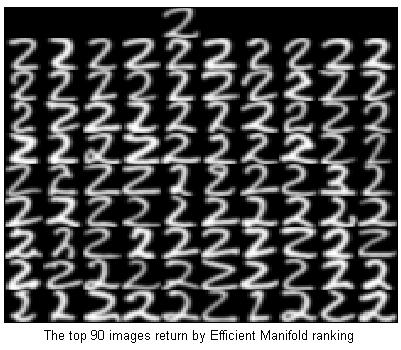
%Ranking with Efficient Manifold ranking
tic;
rand('twister',5489);
opts = [];
opts.p = 500;
y = EMR(fea,y0,opts);
toc;
%Elapsed time is 0.561479 seconds.
[dump,idx]=sort(-y);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(fea(queryIdx,:),[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);


%Clustering on COIL20
clear
load('COIL20.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the original space. MIhat: ',num2str(MIhat)]);
%Clustering in the original space. MIhat: 0.7386
tic;
nBasis = nClass;
SCCoptions.ReguParaType = 'SLEP';
SCCoptions.ReguGamma = 0.2;
[B, V] = SCC(fea', nBasis, SCCoptions); %'
CodingTime = toc;
disp(['Sparse Concept Coding with ',num2str(nBasis),' basis vectors. Time: ',num2str(CodingTime)]);
%Sparse Concept Coding with 20 basis vectors. Time: 1.924
rand('twister',5489);
label = litekmeans(V',nClass,'Replicates',20); %'
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the ',num2str(nBasis),'-dim Sparse Concept Coding space, MIhat: ',num2str(MIhat)]);
%Clustering in the 20-dim Sparse Concept Coding space, MIhat: 0.90176
tic;
SCCoptions.ReguParaType = 'LARs';
SCCoptions.Cardi = 2:2:20;
[B, V, cardiCandi] = SCC(fea', nBasis, SCCoptions); %'
CodingTime = toc;
disp(['Sparse Concept Coding with ',num2str(nBasis),' basis vectors. Time: ',num2str(CodingTime)]);
%Sparse Concept Coding with 20 basis vectors. Time: 4.5091
for i = 1:length(cardiCandi)
rand('twister',5489);
label = litekmeans(V{i}',nClass,'Replicates',20); %'
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the ',num2str(nBasis),'-dim Sparse Concept Coding space with cardinality ',num2str(cardiCandi(i)),'. MIhat: ',num2str(MIhat)]);
end
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 2. MIhat: 0.68255
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 4. MIhat: 0.79441
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 6. MIhat: 0.84102
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 8. MIhat: 0.81865
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 10. MIhat: 0.84306
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 12. MIhat: 0.88502
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 14. MIhat: 0.88996
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 16. MIhat: 0.88736
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 18. MIhat: 0.87489
%Clustering in the 20-dim Sparse Concept Coding space with cardinality 20. MIhat: 0.88158
%Clustering on MNIST (70000 samples with 784 dimensions)
clear;
load('Orig.mat');
%Clustering using kmeans
rand('twister',5489)
tic;res=litekmeans(fea,10,'MaxIter',100,'Replicates',10);toc
%Elapsed time is 137.741150 seconds.
res = bestMap(gnd,res);
AC = length(find(gnd == res))/length(gnd)
MIhat = MutualInfo(gnd,res)
%AC: 0.5389
%MIhat: 0.4852
%Clustering using landmark-based spectral clustering
rand('twister',5489)
tic;res = LSC(fea, 10);toc
%Elapsed time is 20.865842 seconds.
res = bestMap(gnd,res);
AC = length(find(gnd == res))/length(gnd)
MIhat = MutualInfo(gnd,res)
%AC: 0.7270
%MIhat: 0.7222
opts.r = 2;
opts.kmMaxIter = 3;
rand('twister',5489)
tic;res = LSC(fea, 10, opts);toc
%Elapsed time is 15.471343 seconds.
res = bestMap(gnd,res);
AC = length(find(gnd == res))/length(gnd)
MIhat = MutualInfo(gnd,res)
%AC: 0.7585
%MIhat: 0.7437
Deng Cai, Xiaofei He, Xiaoyun Wu, Jiawei Han, "Non-negative Matrix Factorization on Manifold ", ICDM 2008.
%Clustering on COIL20
clear;
load('COIL20.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the original space. MIhat: ',num2str(MIhat)]);
%Clustering in the original space. MIhat: 0.7386
%NMF learning
options = [];
options.maxIter = 100;
options.alpha = 0;
%when alpha = 0, GNMF boils down to the ordinary NMF.
rand('twister',5489);
[U,V] = GNMF(fea',nClass,[],options); %'
%Clustering in the NMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the NMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the NMF subspace. MIhat: 0.74361
%GNMF learning
options = [];
options.WeightMode = 'Binary';
W = constructW(fea,options);
options.maxIter = 100;
options.alpha = 100;
rand('twister',5489);
[U,V] = GNMF(fea',nClass,W,options); %'
%Clustering in the GNMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the GNMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the GNMF subspace. MIhat: 0.87599
%Clustering on PIE
clear;
load('PIE_pose27.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the original space. MIhat: ',num2str(MIhat)]);
%Clustering in the original space. MIhat: 0.5377
%NMF learning
options = [];
options.maxIter = 100;
options.alpha = 0;
%when alpha = 0, GNMF boils down to the ordinary NMF.
rand('twister',5489);
[U,V] = GNMF(fea',nClass,[],options); %'
%Clustering in the NMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the NMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the NMF subspace. MIhat: 0.69824
%GNMF learning
options = [];
options.WeightMode = 'Binary';
W = constructW(fea,options);
options.maxIter = 100;
options.alpha = 100;
rand('twister',5489);
[U,V] = GNMF(fea',nClass,W,options); %'
%Clustering in the GNMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the GNMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the GNMF subspace. MIhat: 0.88074
%Clustering on TDT2
clear;
load('TDT2.mat');
nClass = length(unique(gnd));
%tfidf weighting and normalization
fea = tfidf(fea);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the original space. MIhat: ',num2str(MIhat)]);
%Clustering in the original space. MIhat: 0.75496
%NMF learning
options = [];
options.maxIter = 100;
options.alpha = 0;
%when alpha = 0, GNMF boils down to the ordinary NMF.
rand('twister',5489);
[U,V] = GNMF(fea',nClass,[],options); %'
%Clustering in the NMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the NMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the NMF subspace. MIhat: 0.65132
%GNMF learning
options = [];
options.WeightMode = 'Binary';
W = constructW(fea,options);
options.maxIter = 100;
options.alpha = 100;
rand('twister',5489);
[U,V] = GNMF(fea',nClass,W,options); %'
%Clustering in the GNMF subspace
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the GNMF subspace. MIhat: ',num2str(MIhat)]);
%Clustering in the GNMF subspace. MIhat: 0.8621
%Clustering on TDT2
clear;
load('TDT2_all.mat');
%Download random cluster index file 10Class.rar and uncompress.
%tfidf weighting and normalization
fea = tfidf(fea);
%--------------------------------------
nTotal = 50;
MIhat_KM = zeros(nTotal,1);
MIhat_LCCF = zeros(nTotal,1);
for i = 1:nTotal
load(['top56/10Class/',num2str(i),'.mat']);
feaSet = fea(sampleIdx,:);
feaSet(:,zeroIdx)=[];
gndSet = gnd(sampleIdx);
nClass = length(unique(gndSet));
rand('twister',5489);
label = kmeans(feaSet,nClass,'Distance','cosine','EmptyAction','singleton','Start','cluster','Replicates',10);
label = bestMap(gndSet,label);
MIhat_KM(i) = MutualInfo(gndSet,label);
LCCFoptions = [];
LCCFoptions.WeightMode = 'Cosine';
LCCFoptions.bNormalized = 1;
W = constructW(feaSet,LCCFoptions);
LCCFoptions.maxIter = 200;
LCCFoptions.alpha = 100;
LCCFoptions.KernelType = 'Linear';
LCCFoptions.weight = 'NCW';
rand('twister',5489);
[U,V] = LCCF(feaSet',nClass,W,LCCFoptions); %'
rand('twister',5489);
label = kmeans(V,nClass,'EmptyAction','singleton','Start','cluster','Replicates',10);
label = bestMap(gndSet,label);
MIhat_LCCF(i) = MutualInfo(gndSet,label);
disp([num2str(i),' subset done.']);
end
disp(['Clustering in the original space. MIhat: ',num2str(mean(MIhat_KM))]);
disp(['Clustering in the LCCF subspace. MIhat: ',num2str(mean(MIhat_LCCF))]);
%Clustering in the original space. MIhat: 0.65129
%Clustering in the LCCF subspace. MIhat: 0.9156
Deng Cai, Xiaofei He, Jiawei Han, "Efficient Kernel Discriminant Analysis via Spectral Regression", ICDM 2007.
Code download
See more examples on semi-supervised classification
%Face recognition on PIE
clear;
load('random.mat');
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
options = [];
options.KernelType = 'Gaussian';
options.t = 1;
options.ReguAlpha = 0.001;
for nTrain = [2000 3000 4000 5000 6000 7000 8000]
tic;
model = SRKDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
TimeTrain = toc;
tic;
accuracy = SRKDApredict(fea(8001:end,:), gnd(8001:end), model);
TimeTest = toc;
disp(['SRKDA,',num2str(nTrain),' Training, Errorrate: ',num2str(1-accuracy),' TrainTime: ',num2str(TimeTrain),' TestTime: ',num2str(TimeTest)]);
end
%SRKDA,2000 Training, Errorrate: 0.048115 TrainTime: 0.52305 TestTime: 0.49549
%SRKDA,3000 Training, Errorrate: 0.039392 TrainTime: 0.71913 TestTime: 0.55529
%SRKDA,4000 Training, Errorrate: 0.032358 TrainTime: 1.2583 TestTime: 0.66468
%SRKDA,5000 Training, Errorrate: 0.028981 TrainTime: 2.1578 TestTime: 0.99834
%SRKDA,6000 Training, Errorrate: 0.025324 TrainTime: 3.3049 TestTime: 1.1344
%SRKDA,7000 Training, Errorrate: 0.021947 TrainTime: 4.4947 TestTime: 1.1728
%SRKDA,8000 Training, Errorrate: 0.021666 TrainTime: 6.1601 TestTime: 1.4118
clear;
load('lights=27.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',10);
MIhat = MutualInfo(gnd,label);
disp(['kmeans use all the features. MIhat: ',num2str(MIhat)]);
%kmeans in the original space. MIhat: 0.66064
options.ReducedDim = 64;
eigvector = PCA(fea, options);
newfea = fea*eigvector;
newfea = NormalizeFea(newfea);
%Clustering in 64-dim PCA subspace
rand('twister',5489);
label = litekmeans(newfea,nClass,'Replicates',10);
MIhat = MutualInfo(gnd,label);
disp(['kmeans in the 64-dim PCA subspace. MIhat: ',num2str(MIhat)]);
%kmeans in the 64-dim PCA subspace. MIhat: 0.65946
rand('twister',5489);
W = constructW(fea);
nBasis = 128;
alpha = 1;
beta = 0.1;
nIters = 15;
rand('twister',5489);
warning('off', 'all');
[B, S, stat] = GraphSC(newfea', W, nBasis, alpha, beta, nIters); %'
%Clustering in 128-dim GraphSC subspace
rand('twister',5489);
label = litekmeans(S',nClass,'Replicates',10); %'
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the ',num2str(nBasis),'-dim GraphSC subspace with ',num2str(nIters),' iterations. MIhat: ',num2str(MIhat)]);
%Clustering in the 128-dim GraphSC subspace with 15 iterations. MIhat: 0.80253
nIters = 30;
rand('twister',5489);
warning('off', 'all');
[B, S, stat] = GraphSC(newfea', W, nBasis, alpha, beta, nIters); %'
rand('twister',5489);
label = litekmeans(S',nClass,'Replicates',10); %'
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the ',num2str(nBasis),'-dim GraphSC subspace with ',num2str(nIters),' iterations. MIhat: ',num2str(MIhat)]);
%Clustering in the 128-dim GraphSC subspace with 30 iterations. MIhat: 0.86948
nIters = 100;
rand('twister',5489);
warning('off', 'all');
[B, S, stat] = GraphSC(newfea', W, nBasis, alpha, beta, nIters); %'
rand('twister',5489);
label = litekmeans(S',nClass,'Replicates',10); %'
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the ',num2str(nBasis),'-dim GraphSC subspace with ',num2str(nIters),' iterations. MIhat: ',num2str(MIhat)]);
%Clustering in the 128-dim GraphSC subspace with 100 iterations. MIhat: 0.92865
Code download
See more examples on supervised feature selection using MCFS.
%Unsupervised feature selection on COIL20
clear;
load('COIL20.mat');
%Clustering using all the features
rand('twister',5489);
label = litekmeans(fea,20,'Replicates',20);
MIhat = MutualInfo(gnd,label)
disp(['Clustering using all the ',num2str(size(fea,2)),' features. Clustering MIhat: ',num2str(MIhat)]);
%Clustering using all the 1024 features. Clustering MIhat: 0.76056
%Unsupervised feature selection using MCFS
options = [];
options.nUseEigenfunction = 19;
FeaNumCandi = [10:10:100, 120:20:200];
[FeaIndex,FeaNumCandi] = MCFS_p(fea,FeaNumCandi,options);
%Clustering using selected features
for i = 1:length(FeaNumCandi)
SelectFeaIdx = FeaIndex{i};
feaNew = fea(:,SelectFeaIdx);
rand('twister',5489);
label = litekmeans(feaNew,20,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Selected feature num: ',num2str(FeaNumCandi(i)),', Clustering MIhat: ',num2str(MIhat)]);
end
%Selected feature num: 10, Clustering MIhat: 0.64823
%Selected feature num: 20, Clustering MIhat: 0.71271
%Selected feature num: 30, Clustering MIhat: 0.72217
%Selected feature num: 40, Clustering MIhat: 0.73631
%Selected feature num: 50, Clustering MIhat: 0.74794
%Selected feature num: 60, Clustering MIhat: 0.73676
%Selected feature num: 70, Clustering MIhat: 0.75422
%Selected feature num: 80, Clustering MIhat: 0.76514
%Selected feature num: 90, Clustering MIhat: 0.74173
%Selected feature num: 100, Clustering MIhat: 0.74882
%Selected feature num: 120, Clustering MIhat: 0.75249
%Selected feature num: 140, Clustering MIhat: 0.72999
%Selected feature num: 160, Clustering MIhat: 0.74366
%Selected feature num: 180, Clustering MIhat: 0.74058
%Selected feature num: 200, Clustering MIhat: 0.74729
options.nUseEigenfunction = 1;
FeaIndex = MCFS_p(fea,FeaNumCandi,options);
for i = 1:length(FeaNumCandi)
SelectFeaIdx = FeaIndex{i};
feaNew = fea(:,SelectFeaIdx);
rand('twister',5489);
label = litekmeans(feaNew,20,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Selected feature num: ',num2str(FeaNumCandi(i)),', Clustering MIhat: ',num2str(MIhat)]);
end
%Selected feature num: 10, Clustering MIhat: 0.59015
%Selected feature num: 20, Clustering MIhat: 0.64176
%Selected feature num: 30, Clustering MIhat: 0.71692
%Selected feature num: 40, Clustering MIhat: 0.70853
%Selected feature num: 50, Clustering MIhat: 0.73872
%Selected feature num: 60, Clustering MIhat: 0.75203
%Selected feature num: 70, Clustering MIhat: 0.77577
%Selected feature num: 80, Clustering MIhat: 0.75629
%Selected feature num: 90, Clustering MIhat: 0.7603
%Selected feature num: 100, Clustering MIhat: 0.74933
%Selected feature num: 120, Clustering MIhat: 0.76997
%Selected feature num: 140, Clustering MIhat: 0.74568
%Selected feature num: 160, Clustering MIhat: 0.74576
%Selected feature num: 180, Clustering MIhat: 0.7498
%Selected feature num: 200, Clustering MIhat: 0.73387
options.nUseEigenfunction = 2;
FeaIndex = MCFS_p(fea,FeaNumCandi,options);
for i = 1:length(FeaNumCandi)
SelectFeaIdx = FeaIndex{i};
feaNew = fea(:,SelectFeaIdx);
rand('twister',5489);
label = litekmeans(feaNew,20,'Replicates',20);
MIhat = MutualInfo(gnd,label);
disp(['Selected feature num: ',num2str(FeaNumCandi(i)),', Clustering MIhat: ',num2str(MIhat)]);
end
%Selected feature num: 10, Clustering MIhat: 0.60397
%Selected feature num: 20, Clustering MIhat: 0.65351
%Selected feature num: 30, Clustering MIhat: 0.70341
%Selected feature num: 40, Clustering MIhat: 0.72171
%Selected feature num: 50, Clustering MIhat: 0.75024
%Selected feature num: 60, Clustering MIhat: 0.73423
%Selected feature num: 70, Clustering MIhat: 0.72539
%Selected feature num: 80, Clustering MIhat: 0.73908
%Selected feature num: 90, Clustering MIhat: 0.7155
%Selected feature num: 100, Clustering MIhat: 0.72671
%Selected feature num: 120, Clustering MIhat: 0.74591
%Selected feature num: 140, Clustering MIhat: 0.7646
%Selected feature num: 160, Clustering MIhat: 0.77698
%Selected feature num: 180, Clustering MIhat: 0.77171
%Selected feature num: 200, Clustering MIhat: 0.75548
%Clustering on MNIST test set(10000 samples with 784 dimensions)
clear;
load('Test.mat');
nClass = length(unique(gnd));
options.PCARatio = 0.78;
[eigvector, eigvalue] = PCA(fea, options);
newfea = fea*eigvector;
%Clustering using kmeans
rand('twister',5489)
res=litekmeans(newfea,nClass,'MaxIter',100,'Replicates',10);
res = bestMap(gnd,res);
MIhat = MutualInfo(gnd,res)
disp(['Clustering using kmeans. MIhat: ',num2str(MIhat)]);
%Clustering using kmeans. MIhat: 0.49722
%Clustering using GMM
options.show = 0;
options.lambda = 0;
%LCGMM boils down to the original GMM when lambda=0
rand('twister',5489)
[pkx]= LCGMM(newfea, nClass , [], options);
[dump, clusterIndi] = max(pkx,[],2);
res = bestMap(gnd, clusterIndi);
MIhat = MutualInfo(gnd,res);
disp(['Clustering using GMM. MIhat: ',num2str(MIhat)]);
%Clustering using GMM. MIhat: 0.66932
%Clustering using LCGMM
options.k = 20;
W = constructW(newfea, options);
options.show = 0;
options.lambda = 0.1;
rand('twister',5489)
[pkx]= LCGMM(newfea, nClass , W, options);
[dump, clusterIndi] = max(pkx,[],2);
res = bestMap(gnd, clusterIndi);
MIhat = MutualInfo(gnd,res)
disp(['Clustering using LCGMM. MIhat: ',num2str(MIhat)]);
%Clustering using LCGMM. MIhat: 0.74121
%Clustering using 10 largest categories in TDT2
clear;
load('TDT2_all.mat');
idx = gnd <= 10;
fea = fea(idx,:);
gnd = gnd(idx);
nClass = length(unique(gnd));
%tfidf weighting and normalization
fea = tfidf(fea);
disp('NMF_KL...');
NMFKLoptions = [];
NMFKLoptions.maxIter = 50;
NMFKLoptions.alpha = 0;
%when alpha = 0, GNMF_KL boils down to the ordinary NMF_KL.
NMFKLoptions.weight = 'NCW';
nFactor = 10;
rand('twister',5489);
[U, V] = GNMF_KL(fea', nFactor, [], NMFKLoptions); %'
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',10); %'
label = bestMap(gnd,label);
AC = length(find(gnd == label))/length(gnd);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the NMF_KL space. AC=',num2str(AC),' NMI=',num2str(MIhat)]);
%Clustering in the NMF_KL space. AC=0.76207 NMI=0.76042
disp('GNMF_KL...');
Woptions = [];
Woptions.WeightMode = 'Cosine';
Woptions.k = 7;
W = constructW(fea,Woptions);
GNMFKLoptions = [];
GNMFKLoptions.maxIter = 50;
GNMFKLoptions.alpha = 100;
GNMFKLoptions.weight = 'NCW';
nFactor = 10;
rand('twister',5489);
[U, V] = GNMF_KL(fea', nFactor, W, GNMFKLoptions); %'
rand('twister',5489);
label = litekmeans(V,nClass,'Replicates',10); %'
label = bestMap(gnd,label);
AC = length(find(gnd == label))/length(gnd);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the GNMF_KL space. AC=',num2str(AC),' NMI=',num2str(MIhat)]);
%Clustering in the GNMF_KL space. AC=0.89351 NMI=0.82619
%Clustering using 10 largest categories in TDT2
clear;
load('TDT2_all.mat');
idx = gnd <= 10;
fea = fea(idx,:);
gnd = gnd(idx);
nClass = length(unique(gnd));
disp('PLSA...');
PLSAoptions = [];
PLSAoptions.maxIter = 100;
PLSAoptions.alpha = 0;
%when alpha = 0, LTM boils down to the ordinary PLSA.
nTopics = 50;
rand('twister',5489);
[Pz_d_PLSA, Pw_z_PLSA] = LTM(fea',nTopics,[],PLSAoptions); %'
rand('twister',5489);
label = litekmeans(Pz_d_PLSA',nClass,'Replicates',10); %'
label = bestMap(gnd,label);
AC = length(find(gnd == label))/length(gnd);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the PLSA space. AC=',num2str(AC),' NMI=',num2str(MIhat)]);
%Clustering in the PLSA space. AC=0.40866 NMI=0.23675
disp('LTM...');
Woptions = [];
Woptions.WeightMode = 'Cosine';
Woptions.k = 7;
W = constructW(fea,Woptions);
LTMoptions = [];
LTMoptions.maxIter = 100;
LTMoptions.alpha = 100;
nTopics = 50;
rand('twister',5489);
[Pz_d_LTM, Pw_z_LTM] = LTM(fea',nTopics,W,LTMoptions); %'
rand('twister',5489);
label = litekmeans(Pz_d_LTM',nClass,'Replicates',10); %'
label = bestMap(gnd,label);
AC = length(find(gnd == label))/length(gnd);
MIhat = MutualInfo(gnd,label);
disp(['Clustering in the LTM space. AC=',num2str(AC),' NMI=',num2str(MIhat)]);
%Clustering in the LTM space. AC=0.86159 NMI=0.8257
%Clustering on TDT2
clear;
load('TDT2.mat');
%Download random cluster index file 10Class.rar and uncompress.
%tfidf weighting and normalization
fea = tfidf(fea);
%--------------------------------------
nTotal = 50;
MIhat_KM = zeros(nTotal,1);
MIhat_LapPLSI = zeros(nTotal,1);
for i = 1:nTotal
load(['top30/10Class/',num2str(i),'.mat']);
feaSet = fea(sampleIdx,:);
feaSet(:,zeroIdx)=[];
gndSet = gnd(sampleIdx);
nClass = length(unique(gndSet));
rand('twister',5489);
label = litekmeans(feaSet,nClass,'Replicates',10);
label = bestMap(gndSet,label);
MIhat_KM(i) = MutualInfo(gndSet,label);
LapPLSIoptions = [];
LapPLSIoptions.WeightMode = 'Cosine';
LapPLSIoptions.bNormalized = 1;
LapPLSIoptions.k = 7;
W = constructW(feaSet,LapPLSIoptions);
LapPLSIoptions.maxIter = 100;
LapPLSIoptions.lambda = 1000;
nTopics = 20;
rand('twister',5489);
[Pz_d, Pw_z] = LapPLSI(feaSet', nTopics, W, LapPLSIoptions); %'
rand('twister',5489);
label = litekmeans(Pz_d',nClass,'Replicates',10); %'
label = bestMap(gndSet,label);
MIhat_LapPLSI(i) = MutualInfo(gndSet,label);
disp([num2str(i),' subset done.']);
end
disp(['Clustering in the original space. MIhat: ',num2str(mean(MIhat_KM))]);
disp(['Clustering in the LapPLSI subspace. MIhat: ',num2str(mean(MIhat_LapPLSI))]);
%Clustering in the original space. MIhat: 0.71259
%Clustering in the LapPLSI subspace. MIhat: 0.89509
%Face recognition on PIE
clear;
load('random.mat');
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
options = [];
options.ReguAlpha = 0.1;
for nTrain = [2000 3000 4000 5000 6000 7000 8000]
tic;
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
TimeTrain = toc;
tic;
accuracy = SRDApredict(fea(8001:end,:), gnd(8001:end), model);
TimeTest = toc;
disp(['SRDA,',num2str(nTrain),' Training, Errorrate: ',num2str(1-accuracy),' TrainTime: ',num2str(TimeTrain),' TestTime: ',num2str(TimeTest)]);
end
%SRDA,2000 Training, Errorrate: 0.06359 TrainTime: 0.12366 TestTime: 0.013672
%SRDA,3000 Training, Errorrate: 0.054586 TrainTime: 0.16526 TestTime: 0.014179
%SRDA,4000 Training, Errorrate: 0.046989 TrainTime: 0.17939 TestTime: 0.013864
%SRDA,5000 Training, Errorrate: 0.045582 TrainTime: 0.21096 TestTime: 0.014473
%SRDA,6000 Training, Errorrate: 0.041925 TrainTime: 0.25519 TestTime: 0.014044
%SRDA,7000 Training, Errorrate: 0.039674 TrainTime: 0.33038 TestTime: 0.013682
%SRDA,8000 Training, Errorrate: 0.040518 TrainTime: 0.34291 TestTime: 0.014237
%Face recognition on PIE
clear;
load('PIE_pose27_v2.mat');
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%Generate Train/Test split
nSmp = size(fea,1);
rand('twister',5489);
randIdx = randperm(nSmp);
fea = fea(randIdx,:);
gnd = gnd(randIdx);
%Training SRDA with L2-regularization
nTrainList = [floor(nSmp/3) floor(nSmp/2) floor(2*nSmp/3)];
options = [];
options.ReguType = 'Ridge';
options.ReguAlpha = 0.01;
for nTrain = nTrainList
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
accuracy = SRDApredict(fea(nTrain+1:end,:), gnd(nTrain+1:end), model);
disp(['SRDA,',num2str(nTrain),' Train, NC Errorrate: ',num2str(1-accuracy)]);
feaTrain = SRDAtest(fea(1:nTrain,:), model);
feaTest = SRDAtest(fea(nTrain+1:end,:), model);
gndTrain = gnd(1:nTrain);
gndTest = gnd(nTrain+1:end);
D = EuDist2(feaTest,feaTrain,0);
[dump,idx] = min(D,[],2);
predictlabel = gndTrain(idx);
errorrate = length(find(predictlabel-gndTest))/length(gndTest);
disp(['SRDA,',num2str(nTrain),' Train, NN Errorrate: ',num2str(errorrate)]);
end
%SRDA,1109 Train, NC Errorrate: 0.030631
%SRDA,1109 Train, NN Errorrate: 0.029279
%SRDA,1664 Train, NC Errorrate: 0.024024
%SRDA,1664 Train, NN Errorrate: 0.022222
%SRDA,2219 Train, NC Errorrate: 0.01982
%SRDA,2219 Train, NN Errorrate: 0.01982
%Training SRDA with L1-regularization (use LARs), (Sparse LDA)
options = [];
options.ReguType = 'Lasso';
options.LASSOway = 'LARs';
options.ReguAlpha = 0.001;
options.LassoCardi = 50:10:200;
nTrain = floor(nSmp/3);
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
%Use nearest center classifer
accuracy = SRDApredict(fea(nTrain+1:end,:), gnd(nTrain+1:end), model);
for i = 1:length(model.LassoCardi)
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NC Errorrate: ',num2str(1-accuracy(i))]);
end
%Sparse SRDA,1109 Train, Cardi=50 NC Errorrate: 0.059009
%Sparse SRDA,1109 Train, Cardi=60 NC Errorrate: 0.05045
%Sparse SRDA,1109 Train, Cardi=70 NC Errorrate: 0.045946
%Sparse SRDA,1109 Train, Cardi=80 NC Errorrate: 0.040991
%Sparse SRDA,1109 Train, Cardi=90 NC Errorrate: 0.040541
%Sparse SRDA,1109 Train, Cardi=100 NC Errorrate: 0.037838
%Sparse SRDA,1109 Train, Cardi=110 NC Errorrate: 0.034234
%Sparse SRDA,1109 Train, Cardi=120 NC Errorrate: 0.033784
%Sparse SRDA,1109 Train, Cardi=130 NC Errorrate: 0.033784
%Sparse SRDA,1109 Train, Cardi=140 NC Errorrate: 0.033333
%Sparse SRDA,1109 Train, Cardi=150 NC Errorrate: 0.031081
%Sparse SRDA,1109 Train, Cardi=160 NC Errorrate: 0.030631
%Sparse SRDA,1109 Train, Cardi=170 NC Errorrate: 0.030631
%Sparse SRDA,1109 Train, Cardi=180 NC Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=190 NC Errorrate: 0.028378
%Sparse SRDA,1109 Train, Cardi=200 NC Errorrate: 0.027928
%Use nearest neighbor classifer
feaTrainAll = SRDAtest(fea(1:nTrain,:), model);
feaTestAll = SRDAtest(fea(nTrain+1:end,:), model);
gndTrain = gnd(1:nTrain);
gndTest = gnd(nTrain+1:end);
for i = 1:length(model.LassoCardi)
feaTrain = feaTrainAll{i};
feaTest = feaTestAll{i};
D = EuDist2(feaTest,feaTrain,0);
[dump,idx] = min(D,[],2);
predictlabel = gndTrain(idx);
errorrate = length(find(predictlabel-gndTest))/length(gndTest);
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NN Errorrate: ',num2str(errorrate)]);
end
%Sparse SRDA,1109 Train, Cardi=50 NN Errorrate: 0.031532
%Sparse SRDA,1109 Train, Cardi=60 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=70 NN Errorrate: 0.029279
%Sparse SRDA,1109 Train, Cardi=80 NN Errorrate: 0.03018
%Sparse SRDA,1109 Train, Cardi=90 NN Errorrate: 0.029279
%Sparse SRDA,1109 Train, Cardi=100 NN Errorrate: 0.027928
%Sparse SRDA,1109 Train, Cardi=110 NN Errorrate: 0.028378
%Sparse SRDA,1109 Train, Cardi=120 NN Errorrate: 0.03018
%Sparse SRDA,1109 Train, Cardi=130 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=140 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=150 NN Errorrate: 0.029279
%Sparse SRDA,1109 Train, Cardi=160 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=170 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=180 NN Errorrate: 0.028829
%Sparse SRDA,1109 Train, Cardi=190 NN Errorrate: 0.028378
%Sparse SRDA,1109 Train, Cardi=200 NN Errorrate: 0.028378
nTrain = floor(nSmp/2);
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
%Use nearest center classifer
accuracy = SRDApredict(fea(nTrain+1:end,:), gnd(nTrain+1:end), model);
for i = 1:length(model.LassoCardi)
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NC Errorrate: ',num2str(1-accuracy(i))]);
end
%Sparse SRDA,1664 Train, Cardi=50 NC Errorrate: 0.048649
%Sparse SRDA,1664 Train, Cardi=60 NC Errorrate: 0.042643
%Sparse SRDA,1664 Train, Cardi=70 NC Errorrate: 0.039039
%Sparse SRDA,1664 Train, Cardi=80 NC Errorrate: 0.034234
%Sparse SRDA,1664 Train, Cardi=90 NC Errorrate: 0.030631
%Sparse SRDA,1664 Train, Cardi=100 NC Errorrate: 0.028829
%Sparse SRDA,1664 Train, Cardi=110 NC Errorrate: 0.027027
%Sparse SRDA,1664 Train, Cardi=120 NC Errorrate: 0.025225
%Sparse SRDA,1664 Train, Cardi=130 NC Errorrate: 0.023423
%Sparse SRDA,1664 Train, Cardi=140 NC Errorrate: 0.023423
%Sparse SRDA,1664 Train, Cardi=150 NC Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=160 NC Errorrate: 0.022222
%Sparse SRDA,1664 Train, Cardi=170 NC Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=180 NC Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=190 NC Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=200 NC Errorrate: 0.021622
%Use nearest neighbor classifer
feaTrainAll = SRDAtest(fea(1:nTrain,:), model);
feaTestAll = SRDAtest(fea(nTrain+1:end,:), model);
gndTrain = gnd(1:nTrain);
gndTest = gnd(nTrain+1:end);
for i = 1:length(model.LassoCardi)
feaTrain = feaTrainAll{i};
feaTest = feaTestAll{i};
D = EuDist2(feaTest,feaTrain,0);
[dump,idx] = min(D,[],2);
predictlabel = gndTrain(idx);
errorrate = length(find(predictlabel-gndTest))/length(gndTest);
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NN Errorrate: ',num2str(errorrate)]);
end
%Sparse SRDA,1664 Train, Cardi=50 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=60 NN Errorrate: 0.02042
%Sparse SRDA,1664 Train, Cardi=70 NN Errorrate: 0.02042
%Sparse SRDA,1664 Train, Cardi=80 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=90 NN Errorrate: 0.01982
%Sparse SRDA,1664 Train, Cardi=100 NN Errorrate: 0.019219
%Sparse SRDA,1664 Train, Cardi=110 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=120 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=130 NN Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=140 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=150 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=160 NN Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=170 NN Errorrate: 0.021622
%Sparse SRDA,1664 Train, Cardi=180 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=190 NN Errorrate: 0.021021
%Sparse SRDA,1664 Train, Cardi=200 NN Errorrate: 0.021021
nTrain = floor(2*nSmp/3);
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
%Use nearest center classifer
accuracy = SRDApredict(fea(nTrain+1:end,:), gnd(nTrain+1:end), model);
for i = 1:length(model.LassoCardi)
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NC Errorrate: ',num2str(1-accuracy(i))]);
end
%Sparse SRDA,2219 Train, Cardi=50 NC Errorrate: 0.041441
%Sparse SRDA,2219 Train, Cardi=60 NC Errorrate: 0.033333
%Sparse SRDA,2219 Train, Cardi=70 NC Errorrate: 0.028829
%Sparse SRDA,2219 Train, Cardi=80 NC Errorrate: 0.028829
%Sparse SRDA,2219 Train, Cardi=90 NC Errorrate: 0.028829
%Sparse SRDA,2219 Train, Cardi=100 NC Errorrate: 0.028829
%Sparse SRDA,2219 Train, Cardi=110 NC Errorrate: 0.021622
%Sparse SRDA,2219 Train, Cardi=120 NC Errorrate: 0.018919
%Sparse SRDA,2219 Train, Cardi=130 NC Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=140 NC Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=150 NC Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=160 NC Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=170 NC Errorrate: 0.018919
%Sparse SRDA,2219 Train, Cardi=180 NC Errorrate: 0.018919
%Sparse SRDA,2219 Train, Cardi=190 NC Errorrate: 0.018919
%Sparse SRDA,2219 Train, Cardi=200 NC Errorrate: 0.018919
%Use nearest neighbor classifer
feaTrainAll = SRDAtest(fea(1:nTrain,:), model);
feaTestAll = SRDAtest(fea(nTrain+1:end,:), model);
gndTrain = gnd(1:nTrain);
gndTest = gnd(nTrain+1:end);
for i = 1:length(model.LassoCardi)
feaTrain = feaTrainAll{i};
feaTest = feaTestAll{i};
D = EuDist2(feaTest,feaTrain,0);
[dump,idx] = min(D,[],2);
predictlabel = gndTrain(idx);
errorrate = length(find(predictlabel-gndTest))/length(gndTest);
disp(['Sparse SRDA,',num2str(nTrain),' Train, Cardi=',num2str(model.LassoCardi(i)),' NN Errorrate: ',num2str(errorrate)]);
end
%Sparse SRDA,2219 Train, Cardi=50 NN Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=60 NN Errorrate: 0.016216
%Sparse SRDA,2219 Train, Cardi=70 NN Errorrate: 0.016216
%Sparse SRDA,2219 Train, Cardi=80 NN Errorrate: 0.016216
%Sparse SRDA,2219 Train, Cardi=90 NN Errorrate: 0.017117
%Sparse SRDA,2219 Train, Cardi=100 NN Errorrate: 0.017117
%Sparse SRDA,2219 Train, Cardi=110 NN Errorrate: 0.016216
%Sparse SRDA,2219 Train, Cardi=120 NN Errorrate: 0.016216
%Sparse SRDA,2219 Train, Cardi=130 NN Errorrate: 0.017117
%Sparse SRDA,2219 Train, Cardi=140 NN Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=150 NN Errorrate: 0.017117
%Sparse SRDA,2219 Train, Cardi=160 NN Errorrate: 0.017117
%Sparse SRDA,2219 Train, Cardi=170 NN Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=180 NN Errorrate: 0.018018
%Sparse SRDA,2219 Train, Cardi=190 NN Errorrate: 0.018919
%Sparse SRDA,2219 Train, Cardi=200 NN Errorrate: 0.018018
Single Training Image Face Recognition Data
%Face clustering on PIE
clear;
load('PIE_pose27_v2.mat');
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
%kmeans clustering in the original space
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',10,'Distance','cosine');
MIhat = MutualInfo(gnd,label)
%MIhat: 0.3941
%kmeans in the Laplacian Eigenmap subspace (Spectral Clustering)
rand('twister',5489);
W = constructW(fea);
Y = Eigenmap(W,nClass);
rand('twister',5489);
labelNew = litekmeans(Y,68,'Replicates',10,'Distance','cosine');
MIhatNew = MutualInfo(gnd,labelNew)
%MIhat: 0.6899
%Sparse Spectral Regression subspace learning (Sparse LPP)
USRoptions = [];
USRoptions.W = W;
USRoptions.ReguType = 'Lasso';
USRoptions.LASSOway = 'LARs';
USRoptions.ReguAlpha = 0.01;
USRoptions.ReducedDim = nClass;
USRoptions.LassoCardi = 50:10:250;
USRoptions.bCenter = 0;
model = USRtrain(fea, USRoptions);
feaNew = USRtest(fea, nClass, model);
%kmeans in the Sparse Spectral Regression subspace (Sparse LPP)
for i = 1:length(model.LassoCardi)
rand('twister',5489);
labelNew = litekmeans(feaNew{i},nClass,'Replicates',10,'Distance','cosine');
MIhatNew = MutualInfo(gnd,labelNew);
disp(['Sparse SR subspace, Cardi=',num2str(model.LassoCardi(i)),', MIhat: ',num2str(MIhatNew)]);
end
%Sparse SR subspace, Cardi=50, MIhat: 0.73313
%Sparse SR subspace, Cardi=60, MIhat: 0.76012
%Sparse SR subspace, Cardi=70, MIhat: 0.76716
%Sparse SR subspace, Cardi=80, MIhat: 0.76632
%Sparse SR subspace, Cardi=90, MIhat: 0.77407
%Sparse SR subspace, Cardi=100, MIhat: 0.77838
%Sparse SR subspace, Cardi=110, MIhat: 0.7972
%Sparse SR subspace, Cardi=120, MIhat: 0.78407
%Sparse SR subspace, Cardi=130, MIhat: 0.78408
%Sparse SR subspace, Cardi=140, MIhat: 0.78848
%Sparse SR subspace, Cardi=150, MIhat: 0.78051
%Sparse SR subspace, Cardi=160, MIhat: 0.77967
%Sparse SR subspace, Cardi=170, MIhat: 0.78357
%Sparse SR subspace, Cardi=180, MIhat: 0.77697
%Sparse SR subspace, Cardi=190, MIhat: 0.79725
%Sparse SR subspace, Cardi=200, MIhat: 0.78014
%Sparse SR subspace, Cardi=210, MIhat: 0.79001
%Sparse SR subspace, Cardi=220, MIhat: 0.76642
%Sparse SR subspace, Cardi=230, MIhat: 0.77602
%Sparse SR subspace, Cardi=240, MIhat: 0.77618
%Sparse SR subspace, Cardi=250, MIhat: 0.77628
PIE_pose27.mat contains the data, fea and gnd
TrainTest.mat contains the train/test split. trainIdx, testIdx
1Label folder contains the 20 random label selections. LableIdx, UnlabelIdx
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
feaTrain = fea(trainIdx, :);
gndTrain = gnd(trainIdx);
options = [];
options.Metric = 'Cosine';
options.NeighborMode = 'KNN';
options.WeightMode = 'Cosine';
options.k = 2;
options.bSelfConnected = 0;
W = constructW(feaTrain,options);
options = [];
options.W = W;
options.ReguBeta = 1;
options.ReguAlpha = 0.1;
semisplit = false(size(trainIdx));
semisplit(LabelIdx) = true;
[eigvector] = SDA(gndTrain, feaTrain, semisplit, options);
newfea = fea*eigvector;
Deng Cai, Xiaofei He, and Jiawei Han, "Spectral Regression for Efficient Regularized Subspace Learning", in ICCV'07, 2007. ( pdf )
fea = NormalizeFea(fea);
options = [];
options.NeighborMode = 'KNN';
options.WeightMode = 'Cosine';
options.k = 5;
W = constructW(fea,options);
options = [];
options.W = W;
options.ReguType = 'Ridge';
options.ReguAlpha = 0.1;
[eigvector] = SR_caller(options, fea);
[nSmp,nFea] = size(fea);
if size(eigvector,1) == nFea + 1
newfea = [fea ones(nSmp,1)]*eigvector;
else
newfea = fea*eigvector;
end
%=========Supervised=====================
fea = NormalizeFea(fea);
fea_Train = fea(trainIdx,:);
gnd_Train = gnd(trainIdx);
options = [];
options.gnd = gnd_Train;
options.ReguType = 'Ridge';
options.ReguAlpha = 0.1;
[eigvector] = SR_caller(options, fea_Train);
[nSmp,nFea] = size(fea);
if size(eigvector,1) == nFea + 1
newfea = [fea ones(nSmp,1)]*eigvector;
else
newfea = fea*eigvector;
end
fea = NormalizeFea(fea);
fea_Train = fea(trainIdx,:);
gnd_Train = gnd(trainIdx);
options = [];
options.NeighborMode = 'Supervised';
options.WeightMode = 'Cosine';
options.gnd = gnd_Train;
W = constructW(fea_Train,options);
options.Regu = 1;
options.ReguAlpha = 0.1;
options.ReguType = 'Custom';
load('TensorR_32x32.mat');
options.regularizerR = regularizerR;
[eigvector, eigvalue] = LPP(W, options, fea_Train);
[eigvector, eigvalue] = LDA(gnd_Train, options, fea_Train);
...
newfea = fea*eigvector;
fea = NormalizeFea(fea);
fea_Train = fea(trainIdx,:);
gnd_Train = gnd(trainIdx);
options = [];
options.NeighborMode = 'Supervised';
options.WeightMode = 'Cosine';
options.gnd = gnd_Train;
W = constructW(fea_Train,options);
options.Regu = 0;
options.PCARatio = 1;
bSuccess = 0;
while ~bSuccess
[eigvector, eigvalue, bSuccess] = OLPP(W, options, fea_Train);
end
newfea = fea*eigvector;
fea = NormalizeFea(fea);
fea_Train = fea(trainIdx,:);
gnd_Train = gnd(trainIdx);
options = [];
options.NeighborMode = 'Supervised';
options.WeightMode = 'Cosine';
options.gnd = gnd_Train;
W = constructW(fea_Train,options);
options.nRepeat = 10;
fea_Train = reshape(fea_Train',32,32,size(fea_Train,1));
[U, V, eigvalue_U, eigvalue_V, posIdx] = TensorLPP(fea_Train, W, options);
[nSmp,nFea] = size(fea);
fea = reshape(fea',32,32,nSmp);
nRow = size(U,2);
nCol = size(V,2);
newfea = zeros(nRow,nCol,nSmp);
for i=1:nSmp
newfea(:,:,i) = U'*fea(:,:,i)*V;
end
newfea = reshape(newfea,nRow*nCol,nSmp)';
newfea = newfea(:,posIdx);
clear;
load('lights=27.mat');
nClass = length(unique(gnd));
%Normalize each data vector to have L2-norm equal to 1
fea = NormalizeFea(fea);
Clustering using all the features
rand('twister',5489);
label = litekmeans(fea,nClass,'Replicates',10);
MIhat = MutualInfo(gnd,label);
disp(['kmeans use all the features. MIhat: ',num2str(MIhat)]);
%kmeans using all the features. MIhat: 0.66064
Feature selection using LaplacianScore
rand('twister',5489);
W = constructW(fea);
Y = LaplacianScore(fea, W);
[dump,idx] = sort(-Y);
Clustering using features selected by LaplacianScore
for feaNum = [10:10:300]
newfea = fea(:,idx(1:feaNum));
newfea = NormalizeFea(newfea);
rand('twister',5489);
label = litekmeans(newfea,nClass,'Replicates',10);
MIhat = MutualInfo(gnd,label);
disp(['kmeans use ',num2str(feaNum),' features selected by LaplacianScore. MIhat: ',num2str(MIhat)]);
end
%kmeans use 10 features selected by LaplacianScore. MIhat: 0.71751
%kmeans use 20 features selected by LaplacianScore. MIhat: 0.80963
%kmeans use 30 features selected by LaplacianScore. MIhat: 0.8469
%kmeans use 40 features selected by LaplacianScore. MIhat: 0.85353
%kmeans use 50 features selected by LaplacianScore. MIhat: 0.85302
%kmeans use 60 features selected by LaplacianScore. MIhat: 0.87032
%kmeans use 70 features selected by LaplacianScore. MIhat: 0.84855
%kmeans use 80 features selected by LaplacianScore. MIhat: 0.85341
%kmeans use 90 features selected by LaplacianScore. MIhat: 0.85176
%kmeans use 100 features selected by LaplacianScore. MIhat: 0.86399
%kmeans use 110 features selected by LaplacianScore. MIhat: 0.8615
%kmeans use 120 features selected by LaplacianScore. MIhat: 0.83491
%kmeans use 130 features selected by LaplacianScore. MIhat: 0.84381
%kmeans use 140 features selected by LaplacianScore. MIhat: 0.86288
%kmeans use 150 features selected by LaplacianScore. MIhat: 0.8314
%kmeans use 160 features selected by LaplacianScore. MIhat: 0.84352
%kmeans use 170 features selected by LaplacianScore. MIhat: 0.84474
%kmeans use 180 features selected by LaplacianScore. MIhat: 0.82322
%kmeans use 190 features selected by LaplacianScore. MIhat: 0.83507
%kmeans use 200 features selected by LaplacianScore. MIhat: 0.82185
%kmeans use 210 features selected by LaplacianScore. MIhat: 0.82531
%kmeans use 220 features selected by LaplacianScore. MIhat: 0.83264
%kmeans use 230 features selected by LaplacianScore. MIhat: 0.81115
%kmeans use 240 features selected by LaplacianScore. MIhat: 0.83137
%kmeans use 250 features selected by LaplacianScore. MIhat: 0.83034
%kmeans use 260 features selected by LaplacianScore. MIhat: 0.82588
%kmeans use 270 features selected by LaplacianScore. MIhat: 0.79668
%kmeans use 280 features selected by LaplacianScore. MIhat: 0.7971
%kmeans use 290 features selected by LaplacianScore. MIhat: 0.80705
%kmeans use 300 features selected by LaplacianScore. MIhat: 0.80729