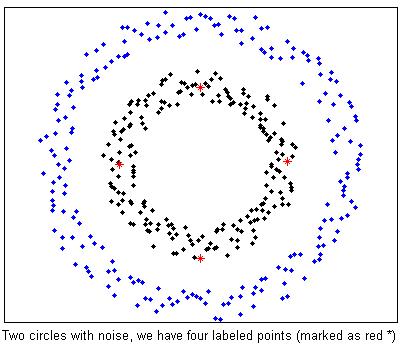
%Two circle data with noise. We already have 4 labeled examples marked as red *
clear;
rand('twister',5489);
[fea, gnd] = GenTwoNoisyCircle();
split = gnd ==1;
figure(1);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
splitLabel = false(length(gnd),1);
splitLabel(1) = true;
splitLabel(50) = true;
splitLabel(100) = true;
splitLabel(150) = true;
hold on;
plot(fea(splitLabel,1),fea(splitLabel,2),'*r');
hold off;
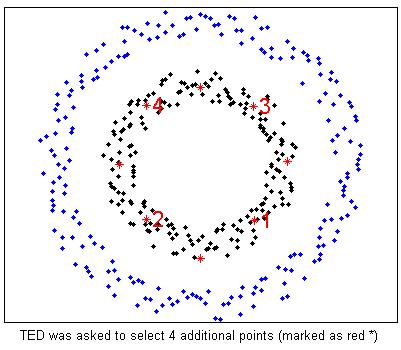
%TED is asked to select 4 more examples (marked as red * and number)
options = [];
options.KernelType = 'Gaussian';
options.t = 0.5;
options.ReguBeta = 0;
%MAED boils down to TED when ReguBeta = 0;
options.splitLabel = splitLabel;
smpRank = MAED(fea,4,options);
figure(2);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
hold on;
plot(fea(splitLabel,1),fea(splitLabel,2),'*r');
for i = 1:length(smpRank)
plot(fea(smpRank(i),1),fea(smpRank(i),2),'*r');
text(fea(smpRank(i),1),fea(smpRank(i),2),['\fontsize{16} \color{red}',num2str(i)]);
end
hold off;
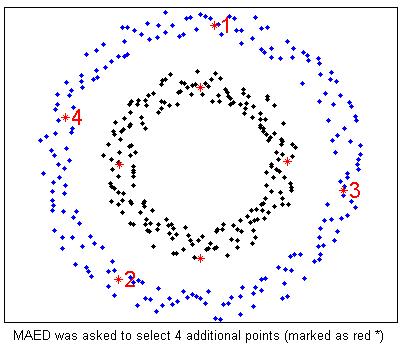
%MAED is asked to select 4 more examples (marked as red * and number)
options = [];
options.KernelType = 'Gaussian';
options.t = 0.5;
options.ReguBeta = 100;
options.splitLabel = splitLabel;
smpRank = MAED(fea,4,options);
figure(3);
plot(fea(split,1),fea(split,2),'.k',fea(~split,1),fea(~split,2),'.b');
hold on;
plot(fea(splitLabel,1),fea(splitLabel,2),'*r');
for i = 1:length(smpRank)
plot(fea(smpRank(i),1),fea(smpRank(i),2),'*r');
text(fea(smpRank(i),1),fea(smpRank(i),2),['\fontsize{16} \color{red}',num2str(i)]);
end
hold off;
%Ranking on the USPS data set (9298 samples with 256 dimensions)
clear;
load('USPS.mat');
gnd = gnd - 1;
%EMR model learning
rand('twister',5489);
opts = [];
opts.p = 500;
[dump, model] = EMR(fea,zeros(size(fea,1),1),opts);
%Generate a query point out of the database.
digit = 2;
idx = find(gnd == digit);
x = mean(fea(idx(1:5),:));
%Ranking with Euclidean distance
D = EuDist2(x,fea);
[dump,idx]=sort(D);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(x,[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);
%Ranking with Efficient Manifold ranking
tic;
[y] = EMRtest(x,model);
toc;
%Elapsed time is 0.015944 seconds.
[dump,idx]=sort(-y);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(x,[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);
%Generate a query point out of the database.
digit = 5;
idx = find(gnd == digit);
x = mean(fea(idx([15,676]),:));
%Ranking with Euclidean distance
D = EuDist2(x,fea);
[dump,idx]=sort(D);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(x,[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);
%Ranking with Efficient Manifold ranking
tic;
[y] = EMRtest(x,model);
toc;
%Elapsed time is 0.015944 seconds.
[dump,idx]=sort(-y);
showfea = fea(idx(2:100),:);
Y = ones(160,160)*-1;
Y(1:16,4*16+1:5*16) = reshape(x,[16,16])'; %'
for i=1:9
for j=0:9
Y(i*16+1:(i+1)*16,j*16+1:(j+1)*16) = reshape(showfea((i-1)*10+j+1,:),[16,16])'; %'
end
end
imagesc(Y);colormap(gray);




%Classification on USPS
load('USPS.mat');
%Classification by SRDA using all the features.
options = [];
options.ReguAlpha = 10;
for nTrain = [2000 3000 4000 5000 6000 7291]
model = SRDAtrain(fea(1:nTrain,:), gnd(1:nTrain), options);
accuracy = SRDApredict(fea(7292:end,:), gnd(7292:end), model);
disp(['SRDA on all ',num2str(size(fea,2)),' features, ',num2str(nTrain),' Train, Errorrate:',num2str(1-accuracy)]);
end
SRDA on all 256 features, 2000 Train, Errorrate:0.12805
SRDA on all 256 features, 3000 Train, Errorrate:0.12207
SRDA on all 256 features, 4000 Train, Errorrate:0.11958
SRDA on all 256 features, 5000 Train, Errorrate:0.12357
SRDA on all 256 features, 6000 Train, Errorrate:0.11908
SRDA on all 256 features, 7291 Train, Errorrate:0.11958
%Classification by SRDA using MCFS selected features.
for nTrain = [2000 3000 4000 5000 6000 7291]
MCFSoptions = [];
MCFSoptions.gnd = gnd(1:nTrain);
FeaNumCandi = [40];
[FeaIndex,FeaNumCandi] = MCFS_p(fea(1:nTrain,:),FeaNumCandi,MCFSoptions);
for i = 1:length(FeaNumCandi)
SelectFeaIdx = FeaIndex{i};
model = SRDAtrain(fea(1:nTrain,SelectFeaIdx), gnd(1:nTrain), options);
accuracy = SRDApredict(fea(8001:end,SelectFeaIdx), gnd(8001:end), model);
disp(['SRDA on selected ',num2str(FeaNumCandi(i)),' features, ',num2str(nTrain),' Train, Errorrate:',num2str(1-accuracy)]);
end
end
SRDA on selected 40 features, 2000 Train, Errorrate:0.12635
SRDA on selected 40 features, 3000 Train, Errorrate:0.13405
SRDA on selected 40 features, 4000 Train, Errorrate:0.13174
SRDA on selected 40 features, 5000 Train, Errorrate:0.13328
SRDA on selected 40 features, 6000 Train, Errorrate:0.14022
SRDA on selected 40 features, 7291 Train, Errorrate:0.13713
%Clustering on COIL20
load('COIL20.mat');
%Clustering in the original space
rand('twister',5489);
label = litekmeans(fea,20,'Replicates',20);
MIhat = MutualInfo(gnd,label)
%MIhat: 0.7606
%SR Learning
USRoptions.ReducedDim = length(unique(gnd))+10;
model = USRtrain(fea, USRoptions);
feaNew = USRtest(fea, length(unique(gnd)), model);
%Clustering in the SR subspace
rand('twister',5489);
labelNew = litekmeans(feaNew,20,'Replicates',20);
MIhatNew = MutualInfo(gnd,labelNew)
%MIhatNew: 0.8974
%Clustering on USPS
load('USPS.mat');
nTrain = 6000;
feaTrain = fea(1:nTrain,:);
gndTrain = gnd(1:nTrain);
feaTest = fea(nTrain+1:end,:);
gndTest = gnd(nTrain+1:end);
%--------------------------------------
%Clustering in the original space
rand('twister',5489);
labelTrain = litekmeans(feaTrain,10,'Replicates',20);
MIhatTrain = MutualInfo(gndTrain,labelTrain)
%MIhatTrain: 0.6343
rand('twister',5489);
labelTest = litekmeans(feaTest,10,'Replicates',20);
MIhatTest = MutualInfo(gndTest,labelTest)
%MIhatTest: 0.5835
%SR Learning
USRoptions.ReducedDim = length(unique(gndTrain))+10;
model = USRtrain(feaTrain, USRoptions);
%Clustering in the SR subspace
feaTrainSR = USRtest(feaTrain, length(unique(gndTrain)), model);
rand('twister',5489);
labelTrainSR = litekmeans(feaTrainSR,10,'Replicates',20);
MIhatTrainSR = MutualInfo(gndTrain,labelTrainSR)
%MIhatTrainSR: 0.7251
feaTestSR = USRtest(feaTest, length(unique(gndTrain)), model);
rand('twister',5489);
labelTestSR = litekmeans(feaTestSR,10,'Replicates',20);
MIhatTestSR = MutualInfo(gndTest,labelTestSR)
%MIhatTestSR: 0.6809
%KSR Learning
UKSRoptions.ReducedDim = length(unique(gndTrain))+10;
model = UKSRtrain(feaTrain, UKSRoptions);
%Clustering in the KSR subspace
feaTrainKSR = UKSRtest(feaTrain, length(unique(gndTrain)), model);
rand('twister',5489);
labelTrainKSR = litekmeans(feaTrainKSR,10,'Replicates',20);
MIhatTrainKSR = MutualInfo(gndTrain,labelTrainKSR)
%MIhatTrainKSR: 0.8195
feaTestKSR = UKSRtest(feaTest, length(unique(gndTrain)), model);
rand('twister',5489);
labelTestKSR = litekmeans(feaTestKSR,10,'Replicates',20);
MIhatTestKSR = MutualInfo(gndTest,labelTestKSR)
%MIhatTestKSR: 0.8070
%Clustering on USPS
load('USPS.mat');
nLabel = 500;
feaLabel = fea(1:nLabel,:);
gndLabel = gnd(1:nLabel);
feaTrain = fea(nLabel+1:7291,:);
feaTest = fea(7292:end,:);
gndTest = gnd(7292:end);
%--------------------------------------
%Training SRKDA on labeled data
options = [];
options.KernelType = 'Gaussian';
options.t = 10;
options.ReguAlpha = 0.01;
model = SRKDAtrain(feaLabel, gndLabel, options);
accuracy = SRKDApredict(feaTest, gndTest, model);
Errorrate = 1-accuracy
%Errorrate: 0.0947
%--------------------------------------
%If we have some additional training data (without label)
options.ReguBeta = 10;
model = SRKDAtrain(feaLabel, gndLabel, options, feaTrain);
accuracy = SRKDApredict(feaTest, gndTest, model);
Errorrate = 1-accuracy
%Errorrate: 0.0708
%If SRKDA can see the test data (without label)
model = SRKDAtrain(feaLabel, gndLabel, options, [feaTrain;feaTest]);
accuracy = SRKDApredict(feaTest, gndTest, model);
Errorrate = 1-accuracy
%Errorrate: 0.0648